CRISPR Point Mutation Stable Cell Line
VectorBuilder can engineer stable cell lines harboring desired point mutations at genomic target sites of interest. These point mutations are generated using a CRISPR-based approach wherein a target site specific gRNA/Cas9 RNP complex is introduced into the target cells along with a donor vector carrying the point mutation template for homology-directed repair (HDR). Our proprietary technology allows us to achieve very efficient gene delivery and HDR, resulting in rapid turnaround and high successful rate for obtaining homozygous mutants.
Highlights
- Supreme point mutation efficiency: cells exhibit HDR rate of up to 50% with our proprietary donor design and gene delivery approach.
- Non-viral delivery approach: electroporation-based RNP delivery is efficient and has minimal off-target effects.
- Rapid turnaround: homozygous mutant cells can be delivered in as fast as 18 weeks starting from vector design.
Service Details
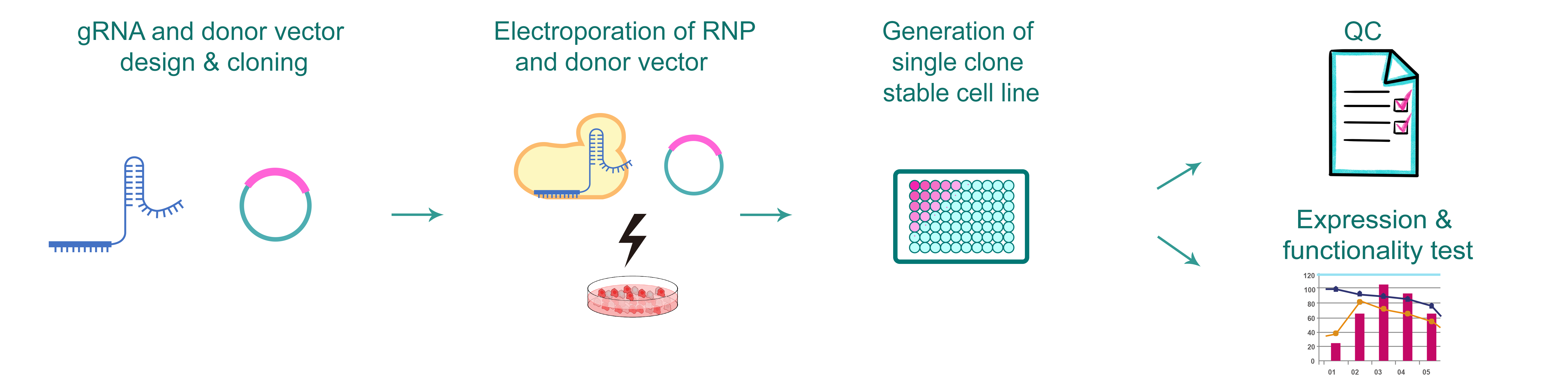
Figure 1. Typical workflow of CRISPR point mutation stable cell line production.
Price and turnaround
| Service Type | Deliverable | Price (USD)* | Turnaround |
|---|---|---|---|
| Point mutation | One homozygous single clone (>106 cells/vial, 2 vials) | From $8,999 | 14-20 weeks |
* Additional charge will apply for extra single clones or vials.
QC assays
| Assay | Methods |
|---|---|
| Point mutation validation (default) | PCR, Sanger sequencing |
| Expression test (add-on) | RT-qPCR, WB, IF, FACS |
| Off-target analysis (add-on) | NGS, PCR, Sanger sequencing |
| Chromosome analysis (add-on) | Karyotyping |
| Sterility (default) | PCR for mycoplasma detection, bioburden test for sterility detection |
Downstream services
We can perform various phenotypic assessments and functional validation of your engineered cell lines, including assays for proliferation, apoptosis, migration, viability, cytotoxicity, etc.
Case Studies
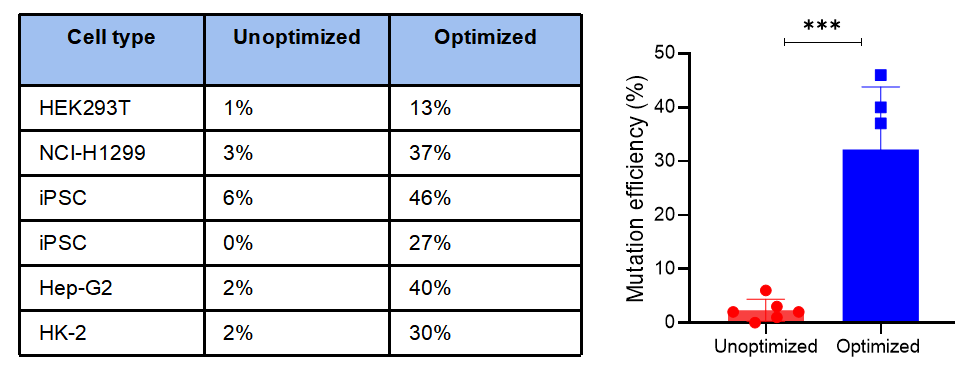
Figure 2. Point mutation rates in various cell types using our optimized mutagenesis strategy.
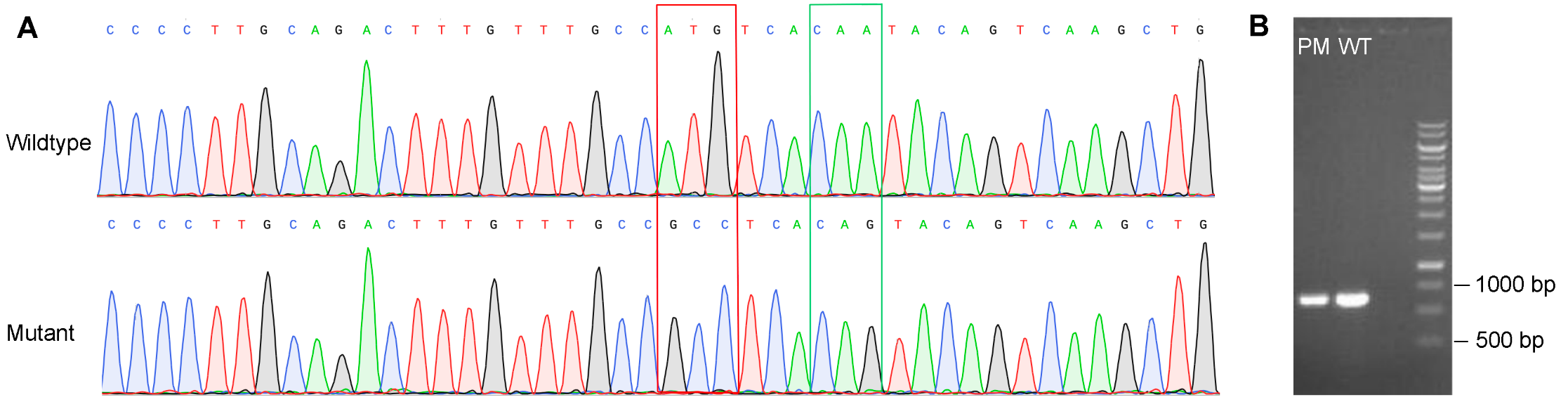
Figure 3. Validation of point mutations in THP-1 cells. (A) Point mutations in the GOI were verified by Sanger sequencing. The red square indicates nonsynonymous mutations and the green square indicates synonymous mutations. (B) Lengths of the PCR product of target site are the same for cells with point mutation (PM) and wildtype (WT) cells, indicating no structural mutation in PM cells.
How to Order
FAQ
For smaller fragments containing the mutation to be introduced, ssODN is a good option because of technical simplicity and its inability to integrate at random locations in the genome. However, for ssODN the template must be less than 200 bp. For larger fragments, dsDNA donors can be used. In either case, efficiency is increased when the cut-to-mutation distance is minimized to 10 base pairs for ssODN and 1 kb for dsDNA donors.
Although synonymous mutations produce the same amino acid sequence, they can affect codon biases, mRNA secondary structure and stability, and protein translation rates. Introducing synonymous mutations in cell lines allows the study of the effects of these post-transcriptional changes. Additionally, introducing a synonymous mutation that disrupts the target sequence for CRISPR components (e.g. PAM and nearby sequences) can suppress unwanted cleavage at off-target and previously CRISPR-Cas9 edited site.



