Barcode Library Construction
VectorBuilder offers state-of-the-art barcode library generation services, enabling researchers to run multiplex analyses at single-cell or single-molecule level during experiments. Your barcode library can be delivered in the format of E. coli stock, plasmid DNA, or recombinant virus. We ensure the uniform and unbiased barcode representation through next-generation sequencing (NGS) validation.
Highlights
- High complexity: We can generate barcode libraries exceeding 108 in complexity.
- High uniformity and unbiased nucleotide distribution
- Full technical support: Our highly experienced scientists can help you optimize barcode library design and choose optimal approach for barcode generation and library cloning.
Service Details
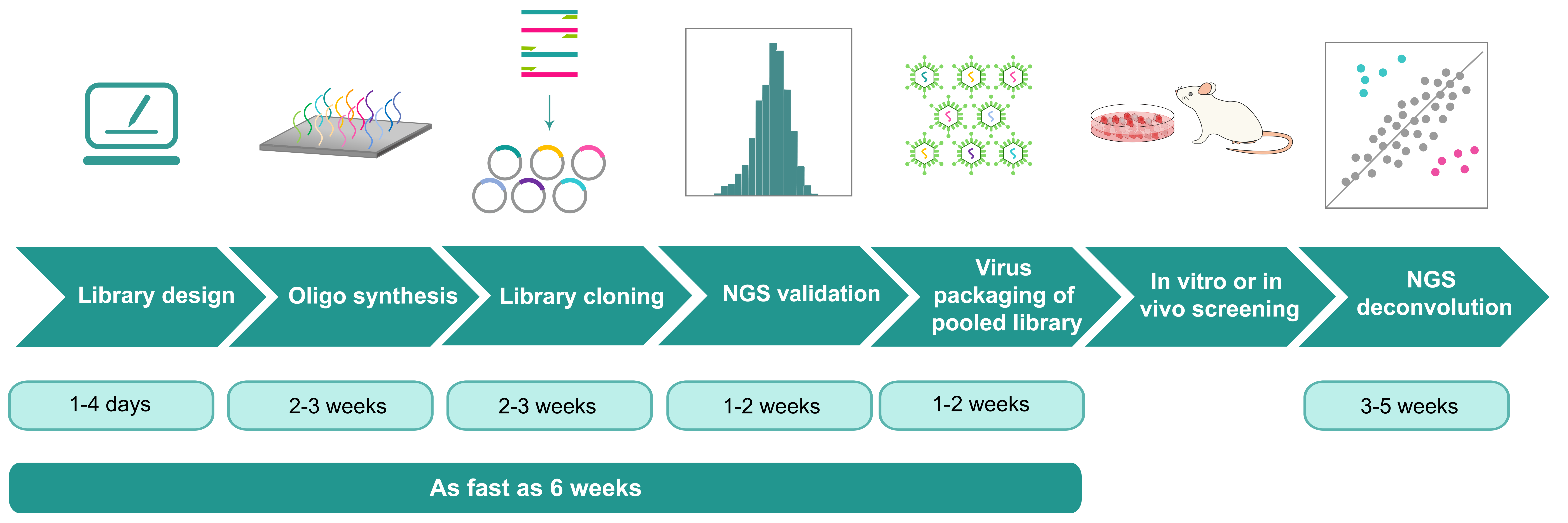
Price and turnaround Price Match
Table 1. Price and turnaround by service module
| Service Module | Brief Description | Price (USD) | Turnaround |
|---|---|---|---|
| Library design | Includes design of barcode sequence, vector backbone, barcode synthesis strategy, and library cloning strategy, to achieve complexity greater than 108, enabling robust and high-throughput applications in genetic and functional studies. | Free | 1-4 days |
| Pooled library cloning | Includes barcode synthesis, massive parallel cloning of barcodes into desired vector backbone, preliminary validation by Sanger sequencing, and full validation by NGS. Deliverable includes library in E. coli glycerol stock and NGS report. See Table 2 below for more details. | ||
| Virus packaging of pooled library | Please click here to view detailed info of virus packaging services. The price for packaging library plasmid is 1.5-fold of the price for packaging single vector plasmid. | ||
| NGS deconvolution of post-screening sample | Includes NGS library preparation from genomic DNA of screened cells, Illumina sequencing (>500x coverage), and data analysis. | From $320 per sample | 3-5 weeks |
Table 2. Library cloning price by library complexity.
| Library Complexity | Price (USD) | Turnaround | |
|---|---|---|---|
| <106 | From $2,800 | 5-8 weeks | |
| 106 ~ 107 | From $4,000 | 5-8 weeks | |
| 107 ~ 108 | From $6,500 | 8-11 weeks | |
| >108 | Please inquire | ||
Technical Information
Barcode length and complexity
Creating a barcode with nucleotides involves assigning unique sequences of DNA bases, to represent different information. To start, a short nucleotide sequence could encode basic information, such as an identifier for a specific cell. For instance, "ATCG" might represent cell 1, while "TAGC" signifies cell 2. As the length of the barcode increases, the complexity and diversity of encoded information expands exponentially. The longer the barcode, the more distinct permutations become available, enhancing the capacity to uniquely identify more variants.
The maximum complexity of a barcode library can be calculated based on the number of possible combinations that can be generated with a given set of nucleotides. For barcodes consisting of randomized nucleotides, there could be four outcomes at each position: A, T, G, or C. The total number of possible combinations (complexity) for a given barcode length (N) is 4N. For example, a randomized NNN barcode has 64 (43) possible combinations.
Barcode vector design
Designing the placement of a barcode in a vector involves strategic considerations to ensure effective readout and compatibility with library cloning, screening, and deconvolution processes. For lentiviral vectors, the barcode should be incorporated within the integrated region of the viral vector, ensuring its stable integration into the host genome along with the associated genetic elements, such as gRNAs or variable regions. Placing the barcode within a transcribed region allows for the possibility of barcode readout with RNA transcripts. To facilitate subsequent analyses, the chosen barcode sequence should be compatible with PCR amplification techniques, allowing for efficient and uniform amplification of the barcode region. Moreover, the barcode design should align with NGS protocols, ensuring detectability and accurate quantification during the sequencing process. Ideally, barcode sequences should not interfere with the studied biological process to avoid artifacts, but this often requires a lot of prior knowledge.
Researchers need to decide between a one-to-one or a multiple-to-one relationship for the barcodes and variable regions. In a one-to-one relationship, a single barcode uniquely represents a variable region, ensuring a straightforward and direct association between barcodes and their respective variable regions. On the other hand, in a multiple-to-one relationship, multiple barcodes can represent the same variable region. This approach provides valuable advantages: firstly, treating these multiple barcodes corresponding to the same variable region as biological replicates enhances statistical power during hit identification. This increased statistical power is particularly beneficial in identifying true positive hits and distinguishing them from random variability in high-throughput experiments. Secondly, the utilization of multiple barcodes for the same variable region enables clonal analysis, which is especially valuable in the study of heterogeneous cell populations, such as tumor cells.
Barcode delivery and NGS readout
To deliver DNA barcodes to cells researchers can use viral vectors such as lentivirus, AAV, retrovirus, and transposon systems such as piggyBac and Sleeping Beauty. The choice of vector depends on factors such as the target cell type, delivery efficiency, and the desired duration of barcode expression. When isolating cell barcodes for NGS analysis, the choice between using RNA or genomic DNA for the readout depends on the delivery system employed to introduce the barcodes into cells. Systems that permanently integrate the barcode into the cellular genomic DNA are conducive to genomic DNA isolation. It is crucial, however, to design the barcodes thoughtfully, ensuring they are amenable to PCR amplification and compatible with NGS sequencing protocols. On the other hand, if the barcode is expressed as part of a transcript, RNA can serve as a readout for the barcodes. In single-cell RNA-sequencing experiments, like Perturb-Seq and CROP-Seq, where both transcriptome and barcode information are captured simultaneously, RNA readout is essential for compatibility with the experimental design.
Experimental data
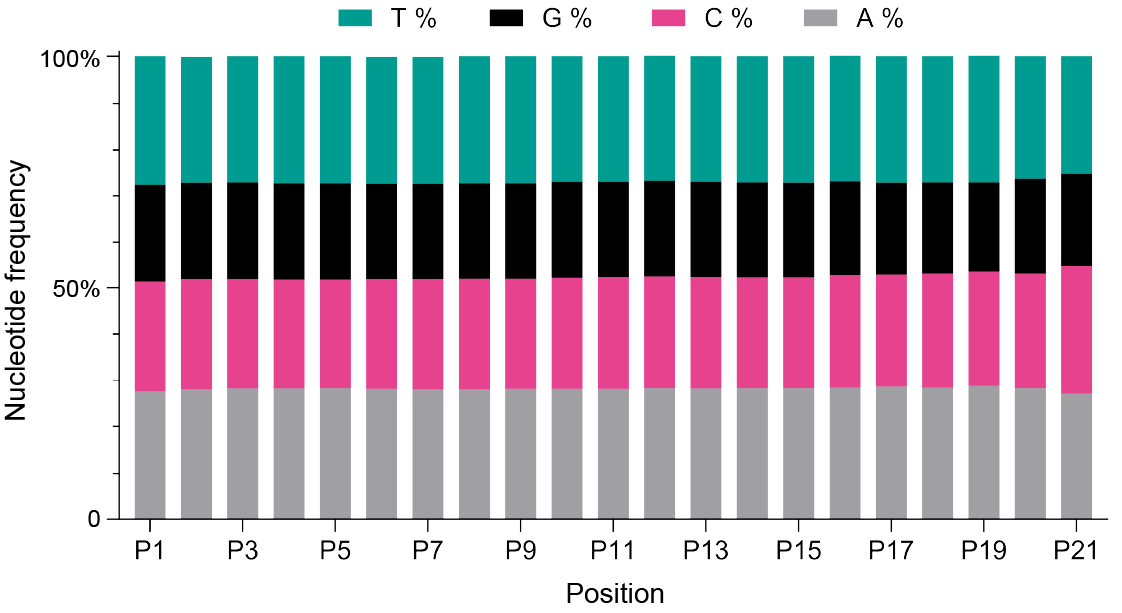
Figure 1. Nucleotide distribution in an N(21) barcode using degenerate nucleotide strategies, illustrating the percentage of adenine (A), cytosine (C), guanine (G), and thymine (T) at each position. The graph reveals an even distribution of nucleotides at all positions.
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.
FAQ
Delivery system: Selection of an appropriate delivery system, such as lentivirus, retrovirus, AAV, or transposon systems, based on the target cells and experimental requirements. Different systems may affect the integration and stability of barcodes in the cellular genome.
Length of barcode: Determining the optimal length of the barcode is crucial. Considerations include the screening scale and the theoretical maximum complexity of the barcode library. Longer barcodes allow for higher complexity but may pose challenges in synthesis and readout.
1-to-1 or multiple-to-1 correlation: Deciding whether each barcode corresponds to a single variable region (1-to-1 correlation) or if multiple barcodes can represent a single variable region. This choice impacts the resolution, specificity, and statistical power of a given library in capturing information.
Barcode sequence design: Choosing the barcode sequence design strategy, which may involve predesigned sequences or random barcode structure (fixed or random). The design affects the synthesis of barcodes and the overall cloning strategy employed in library construction.
Placement of barcode: Carefully considering the placement of barcodes within the genome or transcriptome based on the desired readout method. This includes ensuring compatibility with cloning strategies and accounting for the downstream readout technique, whether it involves sequencing genomic DNA or transcriptomic RNA.
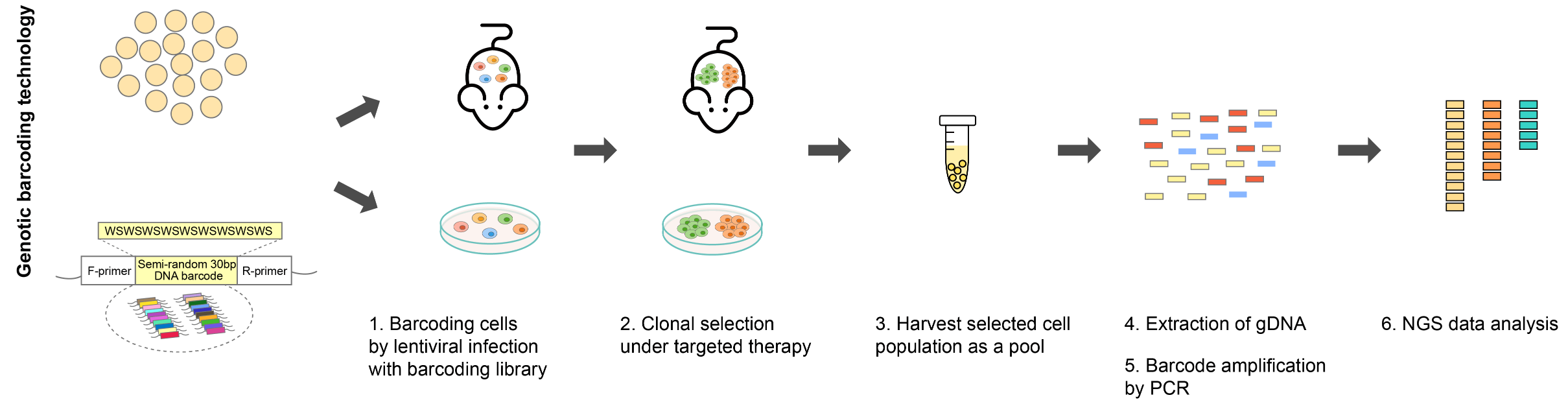
Figure 2. Example workflow of a barcoding experiment.
With viral vector delivery, cell lines of interest are transduced with the barcoding libraries at a low multiplicity of infection (MOI) to label each cell with a unique barcode. The barcoded cell population is then subjected to selective pressure until clonal populations emerge. The harvested resistant clones are pooled, and barcode sequences are PCR-amplified from genomic DNA or isolated from transcripts encoding the barcode information for NGS. The NGS data provides a quantitative readout of the number of clones in the population and the relative abundance of each clone.



