CRISPR Gene Knockout Stable Cell Line
VectorBuilder can engineer stable cell lines with permanent knockout of your gene of interest (GOI). Stable knockout is achieved using a CRISPR-based approach, wherein a single gRNA or a pair of gRNA targeting the GOI is introduced into cells with Cas9 to produce double strand breaks (DSBs). Repair of these breaks typically leads to frameshift mutations or fragment deletions which result in loss-of-function of the target gene.
Highlights
- Full expertise in CRISPR: our team of experts with 10+ years of gene editing experience in vitro and in vivo are capable of designing optimal gene targeting strategy to achieve successful gene knockout.
- Non-viral delivery approach: electroporation-based RNP delivery is efficient, safe, and has minimal off-target effects.
- Rapid turnaround: knockout cells can be produced in as fast as 9 weeks.
Service Details
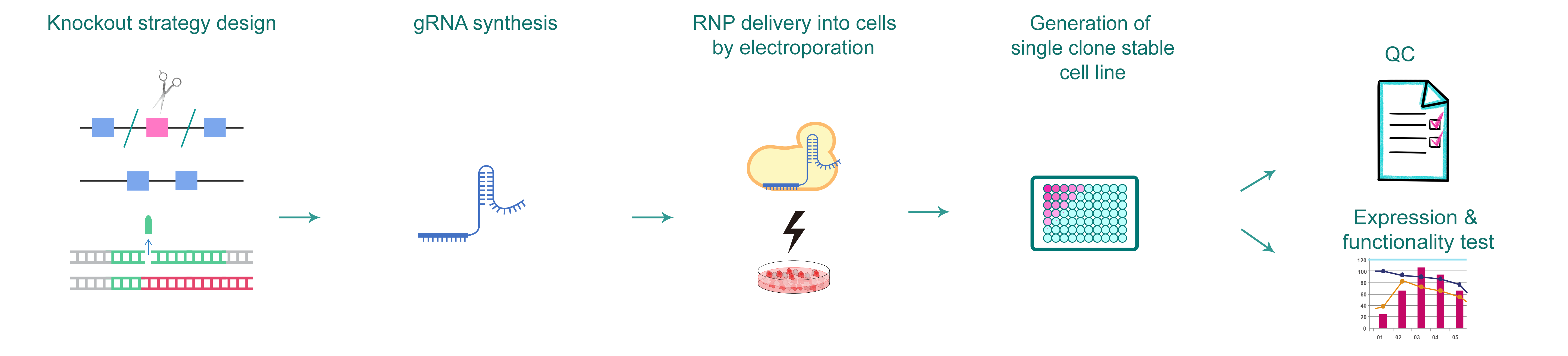
Figure 1. Typical workflow of generating gene knockout cells by CRISPR.
Price and turnaround
| Service Type | Deliverable | Price (USD)* | Turnaround |
|---|---|---|---|
| Knockout (frameshift mutation) | Two homozygous single clones (>106 cells/vial, 2 vials per clone) | From $3,999 | 6-12 weeks |
| Knockout (deletion mutation) | One homozygous single clone (>106 cells/vial, 2 vials) | From $6,999 | 9-15 weeks |
* Additional charge will apply for extra single clones or vials.
QC assays
| Assay | Methods |
|---|---|
| Knockout validation (default) | PCR, Sanger sequencing |
| Expression tests (add-on) | RT-qPCR, WB, IF, FACS |
| Off-target analysis (add-on) | NGS, PCR, Sanger sequencing |
| Chromosome analysis (add-on) | Karyotyping |
| Sterility (default) | PCR for mycoplasma detection, bioburden test for sterility detection |
Downstream services
We can perform various phenotypic assessments and functional validation of your engineered cell lines, including assays for proliferation, apoptosis, migration, viability, cytotoxicity, etc.
Case Studies
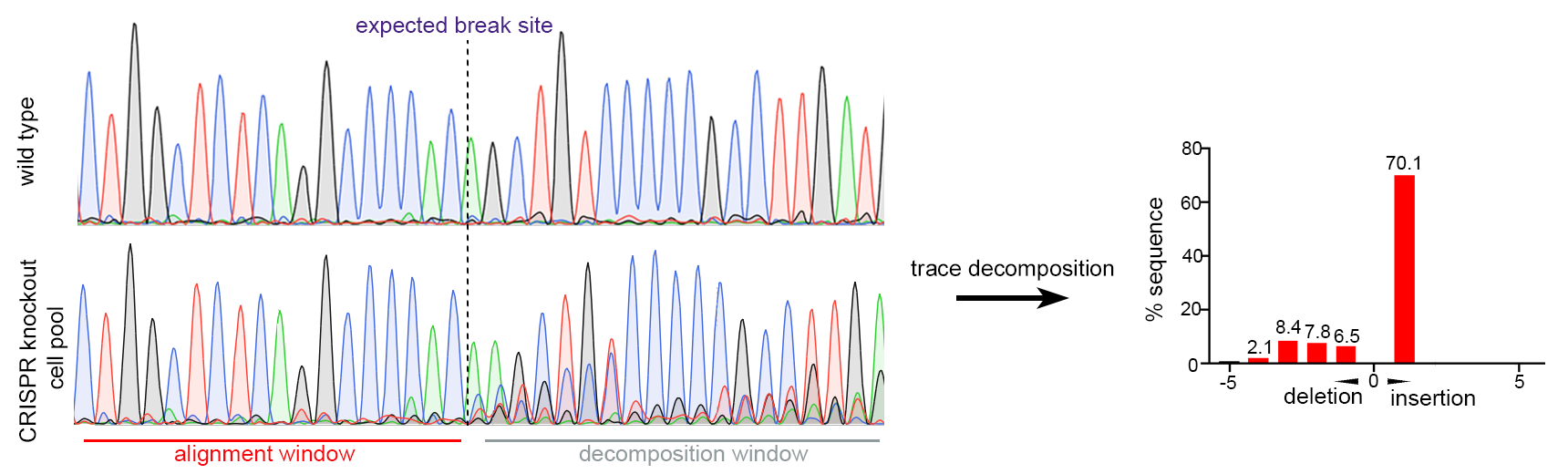
Figure 2. Single gRNA CRISPR-mediated gene knockout in Huh-7 cells. Sanger sequencing of pooled cells showed efficient mutations around CRISPR target site (97.6% of sequence with mutation inferred by trace decomposition).
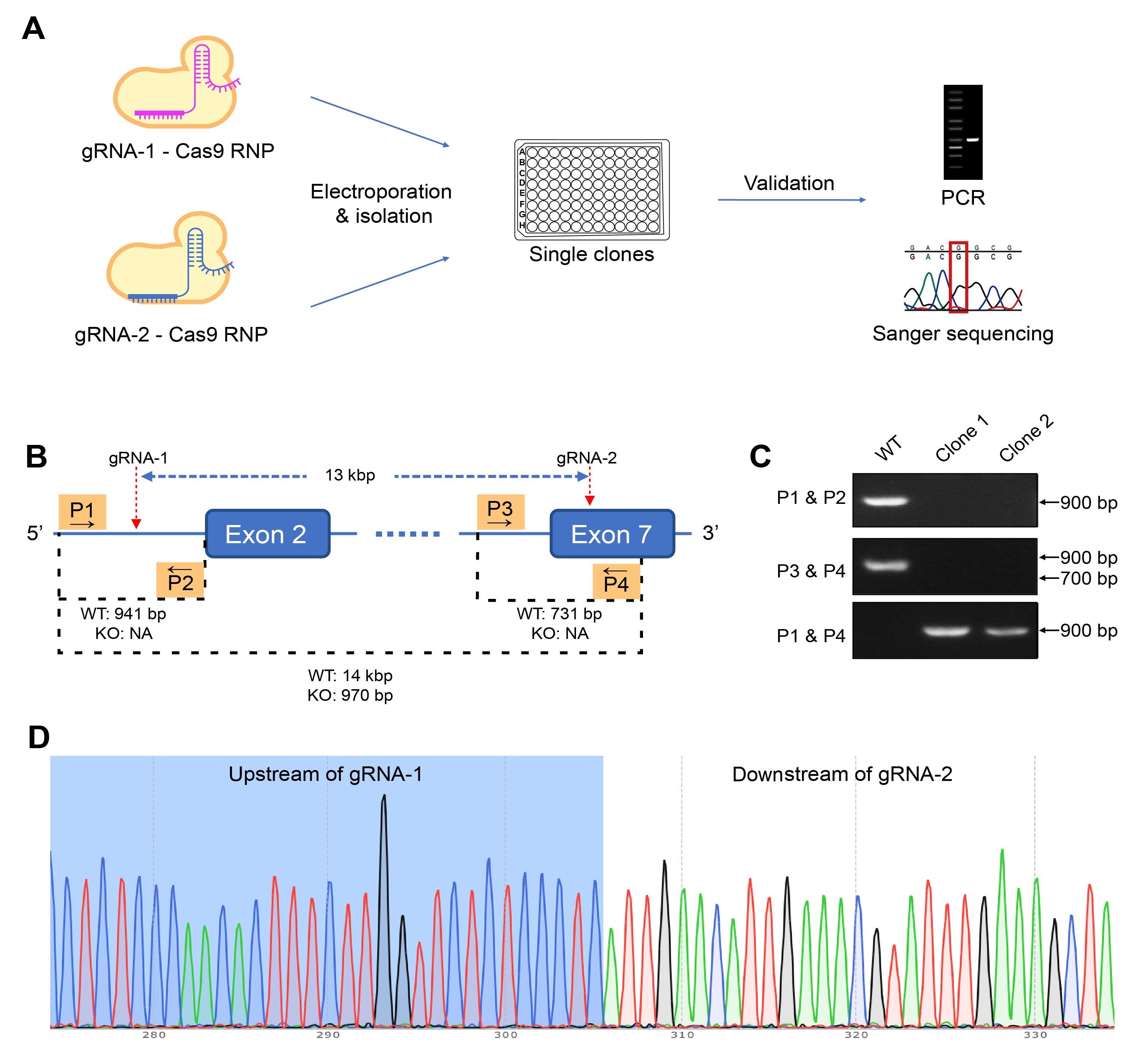
Figure 3. Generating homozygous CD274 knockout (KO) mutants using the gRNA-Cas9 ribonucleoprotein (RNP) approach. (A) The editing RNP is electroporated into target cells, and single clones are isolated and screened. The genotypes of the candidates are validated using PCR and Sanger sequencing. (B) In this case study of editing a murine colon adenocarcinoma cell line, cells were electroporated with RNP binding to two sites on the targeted gene to KO a 13-kbp region. Four primers, P1 to P4, were used in three PCR to differentiate KO and WT clones. Based on the (C) PCR results, clone 1 are validated to be homozygous KO mutants, which is also confirmed by (D) sequencing results.
How to Order
FAQ
Which strategy to employ will depend largely on the target cell and gene. Frameshift mutations using a single gRNA are often sufficient to produce a nonfunctional gene. However, for genes with high levels of compensation, knocking out fragments of multiple genes may be necessary through fragment knockout.
In addition, dual gRNAs can be used if Cas9 nickase is being used to target the two opposite strands of a single target site. In this approach, the nickase enzyme will generate single strand cuts on both strands, one guided by each of the two gRNAs, resulting in DSBs at the target site. Generally, this method reduces off-target effects of CRISPR/Cas9 expression because targeting by both gRNAs is necessary for DSBs to be generated.
In order to decide which method is optimal for your specific application, there are a few things you should consider.
Mechnisms
- Knockdown vectors
Knockdown vectors express short hairpin RNAs (shRNAs) that repress the function of target mRNAs within the cell by inducing their cleavage and repressing their translation. Therefore, shRNA knockdown vectors are not associated with any DNA level sequence change of the gene of interest.
- Knockout vectors
CRISPR functions by directing the Cas9 nuclease to cut specific target sites in the genome. These cuts are then inefficiently repaired by the cellular machinery, resulting in permanent mutations, such as small insertions or deletions, at the sites of repair. A subset of these mutations will result in loss of function of the gene of interest due to frameshifts, premature stop codons, etc. If two closely positioned cut sites in the genome (i.e. within several kb) are targeted simultaneously, this can also result in the deletion of the intervening region.
Effectiveness
shRNA-mediated knockdown will never completely repress the expression of the target gene. Even for the most effective shRNAs, some residual expression of the target gene will remain. In contrast, in a fraction of treated cells, CRISPR can generate permanent mutations which may result in complete loss of gene function.
Consistency and uniformity
shRNA vectors generally provide high cell-to-cell uniformity within the pool of treated cells and very consistent results between experiments. In contrast, CRISPR produces results that are highly non-uniform from cell to cell due to the stochastic nature of the mutations introduced. To fully knock out the GOI in a cell, all copies of the gene in the cell must be knocked out. Given that normal cells have two copies of any gene (except for X- or Y-linked genes) while cancer cells can have more than two copies, such full knockout cells may represent a very small fraction of all the treated cells. For this reason, CRISPR knockout experiments require the screening of clones by sequencing to identify the subset in which all copies of the GOI have been knocked out.
Off-target effects
Off-target effects have been reported for both shRNA-mediated knockdown and CRISPR-mediated knockout. The off-target phenotype(s) can be estimated by using multiple different shRNAs to target the same gene. If a gene knocked down by multiple different shRNAs results in consistent phenotype(s), then it argues against the phenotype(s) being caused by off-target effects. For CRISPR-mediated knockout, multiple clones containing loss-of-function mutations can be analyzed in order to account for any phenotype(s) that may be due to off-target mutations. Additionally, bioinformatically identified off-target sites could be sequenced in the clones to see if they have been mutated.



